PhyloView
I’m excited to share PhyloView, a tool that our team at George Mason University together with David Attaway from ESRI have been working on to bring a new level of insight to understanding how infectious diseases like COVID-19 evolve and spread. PhyloView was developed to harness the vast amounts of phylogenetic and location-based data available on platforms like GISAID, which track genetic sequences of viruses from around the world. Built using ArcGIS Insights, PhyloView allows users to map the transmission paths of a virus over time and across different regions. This work was recently accepted to appear at the 23rd IEEE International Conference on Mobile Data Management (MDM). We will showcase PhyloView’s potential as a valuable tool for public health officials, researchers, and others interested in the complex dynamics of disease ecology.
One of the features I’m particularly excited about is how PhyloView brings multiple types of data together in a single, interactive dashboard. As shown in figure below, users can explore the disease data in several ways: a map that shows geographic spread, a phylogenetic tree that traces viral evolution, and a network visualization of how different strains of the virus are related and spread. These panels are fully interactive, meaning that selecting a case in one panel immediately updates the others. This cross-filtering feature lets users, for instance, zoom into a specific region or a single strain and watch the virus’s spread through that local network over time. Our hope is that this dynamic view will support local health departments in quickly identifying potential transmission hotspots, emerging strains, and important pathways of disease spread.
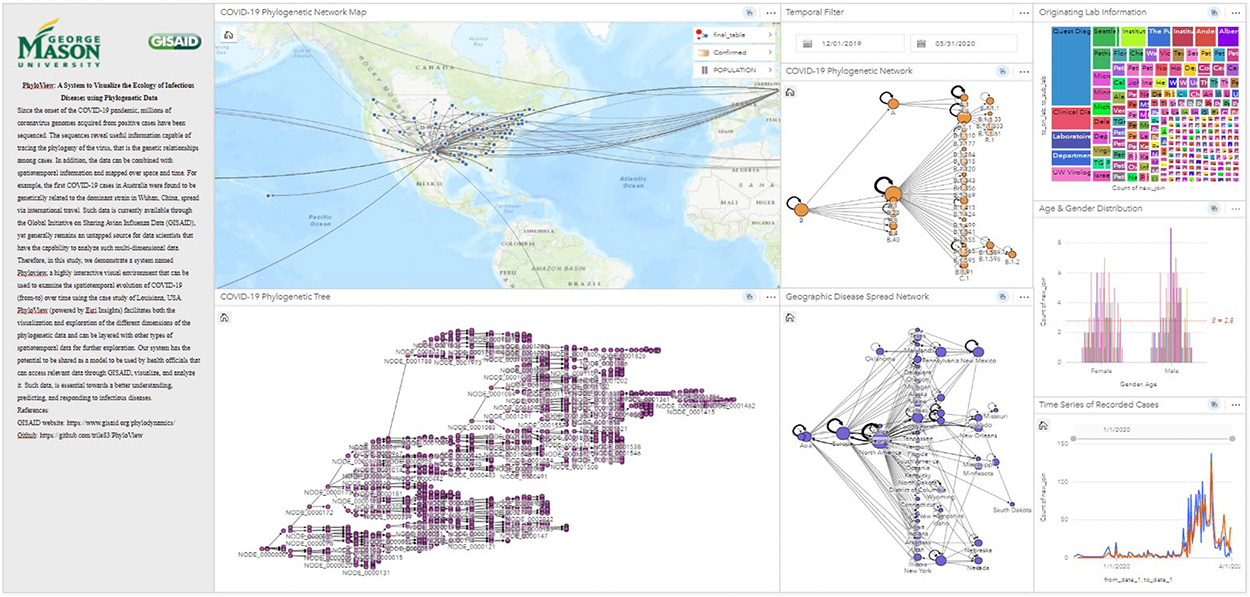
Our aim is that PhyloView will help not only during the current COVID-19 pandemic but also be adaptable to other infectious diseases in the future, allowing us to apply what we’ve learned to better protect communities. One particular tangent we are interested in is to integrate genetic mutations and variants in our data-driven disease models to advance the state-of-the-art.
You can access the paper here.

